Quickview
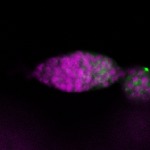
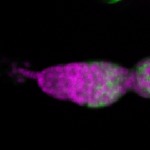
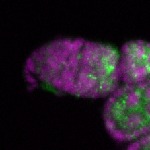
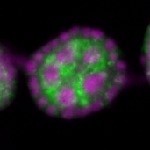
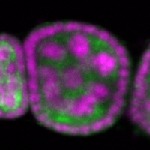
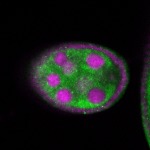
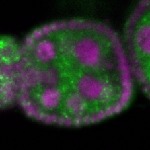
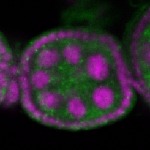
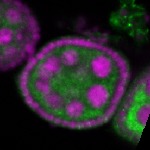
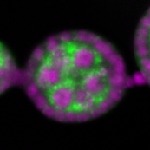
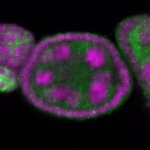
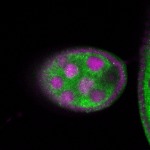
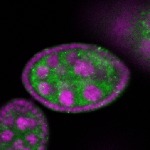
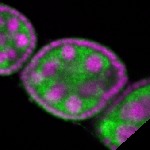
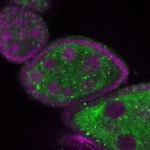
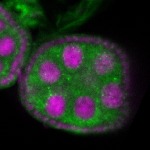
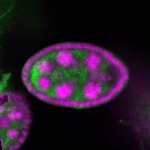
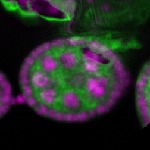
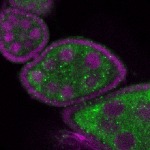
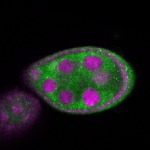
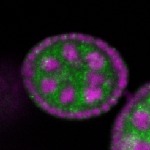
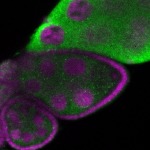
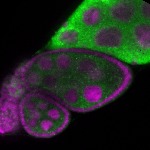
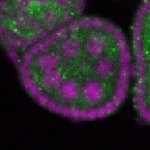
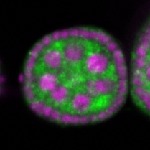
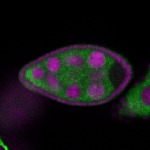
Legend: RNA signal: green, DNA staining: magenta
| Probe source: | LD30817-dg (Flybase link CG6226 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1 | 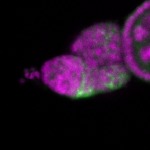 s s
 s s
 s s
 s s | ubiquitous signal at all stages |
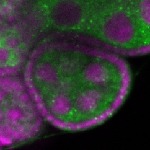
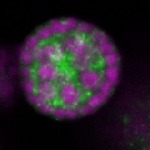
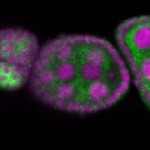
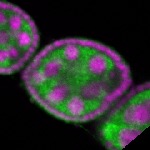
| stage2-7 | 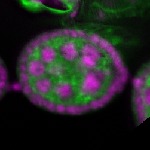 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s | |
| stage8 | 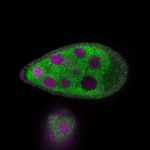 s s
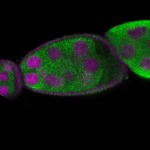 s s
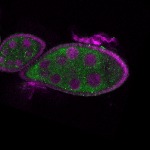 s s | |
| stage9 | 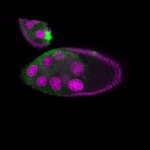 s s
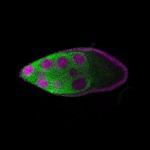 s s
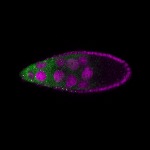 s s | |
| stage10 | 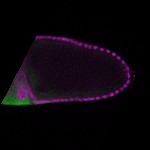 s s
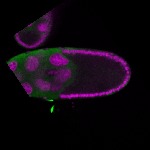 s s
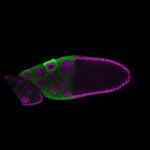 s s
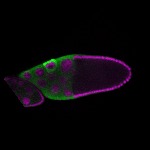 s s | ubiquitous signal |
Quickview
| Probe source: | LD30817 (Flybase link CG6226 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | 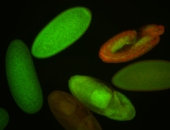
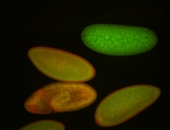
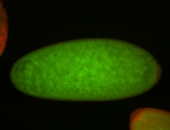
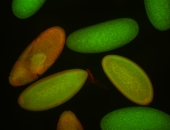
 | Expressed Ubiquitous (Unlocalized) Stage 1-3 Strong expression (1-20ms exp) Maternal |
| stage4-5 | 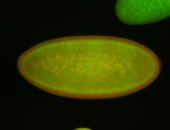
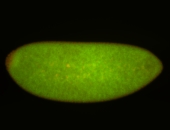
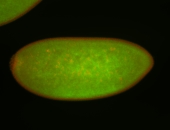 | Pole cell exclusion Stage 4-5 Localized Degraded partially stage 4 Exclusionary patterns Embryonic localization patterns Degradation maternal transcripts Expressed Degraded Maternal |
| stage6-7 | 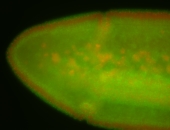
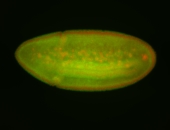
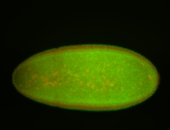
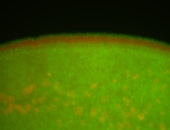
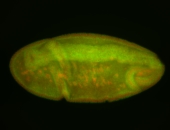
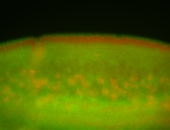 | Stage 6-7 Exclusionary patterns Pole cell exclusion Embryonic localization patterns Expressed Localized Maternal |
| stage8-9 | 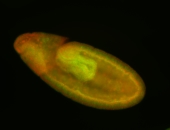
 | Non-expressed Degradation maternal transcripts Degraded Degraded completely stage 9 Stage 8-9 |
Quickview
| Probe source: | LD30817-dg (Flybase link CG6226 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | cat_term:global_finito maternal | |
| stage4-6 | ubiquitous | |
| stage7-8 | ubiquitous trunk mesoderm PR head mesoderm PR ant endoderm A post endoderm PR | |
| stage9-10 | ubiquitous procephalic ectoderm PR trunk mesoderm PR head mesoderm PR ant endoderm PR post endoderm PR | |
| stage11-12 | brain PR dors epidermis PR vent nerve cord PR vent epidermis PR Malpighian tubule PR hindgut proper PR trunk mesoderm PR fat body/gonad PR dors pharyngeal muscle PR post midgut PR ant midgut PR | |
| stage13-16 | cat_term:subset_CNS cat_term:any_neural vent nerve cord anal pad hindgut gonad fat body dors prothoracic pharyngeal muscle midgut gastric caecum | |
| Probe source: | LD30817 (Flybase link CG6226 , FlyFish insitu , BDGP insitu new) | |
| No images or annotation terms | ||