Quickview
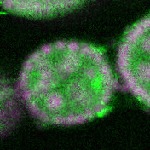
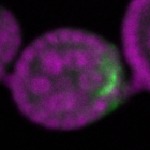
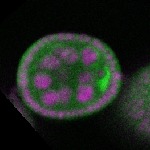
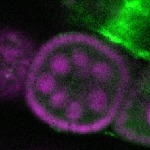
Legend: RNA signal: green, DNA staining: magenta
| Probe source: | RE25591 (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1 |  s s
 s s
 s s | |
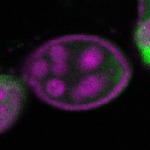
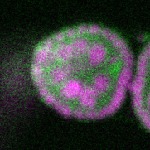
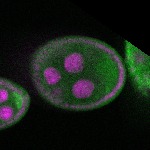
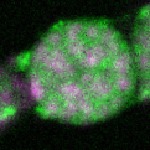
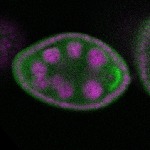
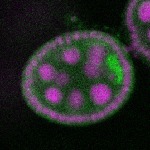
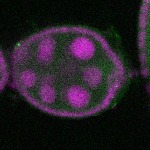
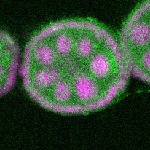
| stage2-7 |  s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
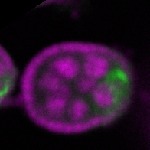 s s
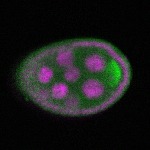 s s
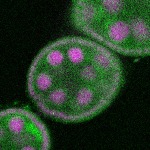 s s
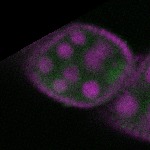 s s
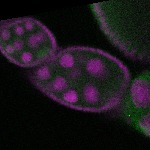 s s
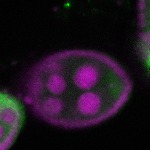 s s | enrichment_ubiquitous oocyte |
| stage8 | 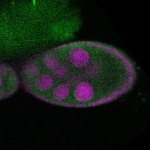 s s
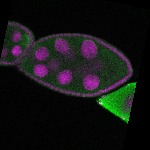 s s | |
| stage9 | 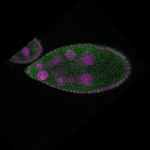 s s
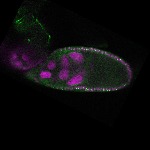 s s
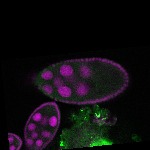 s s | |
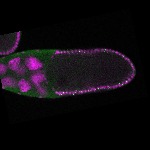
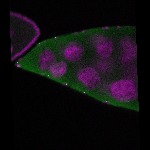
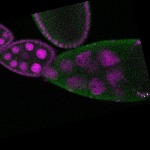
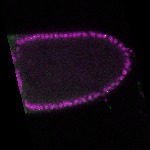
| stage10 | 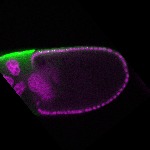 s s
 s s
 s s
 s s
 s s
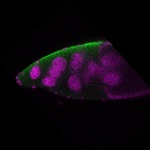 s s | follicle cells overlaying the oocyte nuclear foci somatic cells |
| Probe source: | RE71517 (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
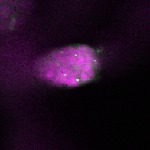
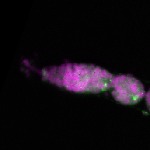
| stage1 | 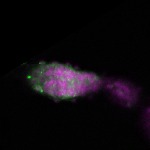 s s
 s s
 s s
 s s | |
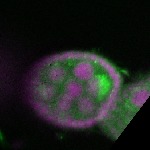
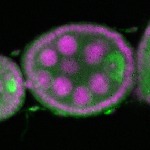
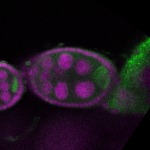
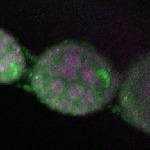
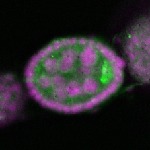
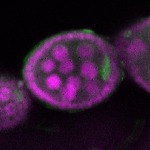
| stage2-7 |  s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
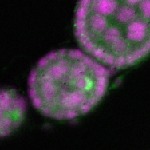 s s | oocyte enrichment_ubiquitous |
| stage8 | 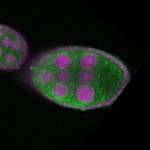 s s
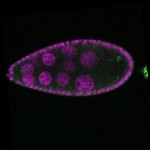 s s | |
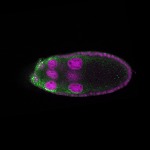
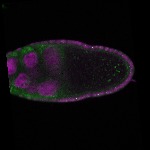
| stage9 | 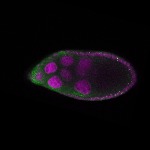 s s
 s s
 s s
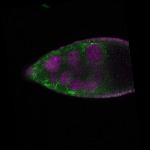 s s
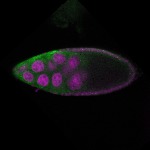 s s | somatic cells follicle cells nuclear foci |
| stage10 | 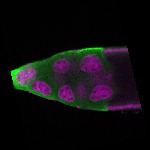 s s
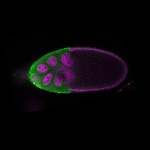 s s
 s s
 s s
 s s | somatic cells follicle cells overlaying the oocyte nuclear foci |
| Probe source: | RE71517-dg (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1 |  s s
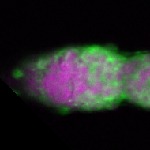 s s
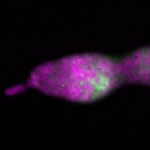 s s
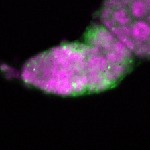 s s
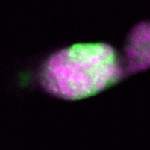 s s | |
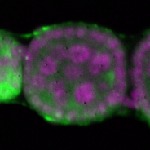
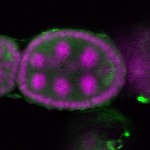
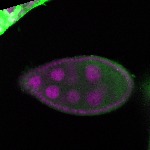
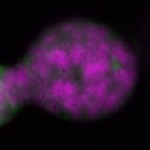
| stage2-7 | 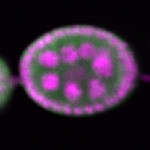 s s
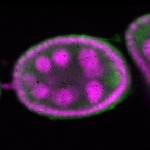 s s
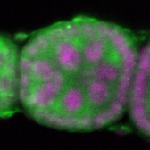 s s
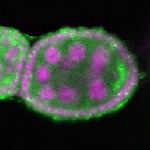 s s
 s s
 s s
 s s
 s s
 s s
 s s | oocyte enrichment_ubiquitous |
| stage8 |  s s
 s s
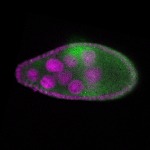 s s | ubiquitous signal |
| stage9 | 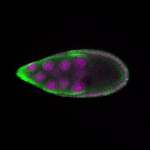 s s
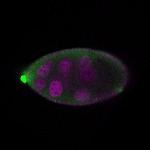 s s
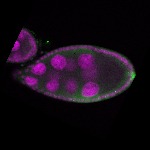 s s | follicle cells nuclear foci germline cells_ubiquitous somatic cells |
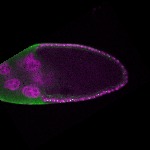
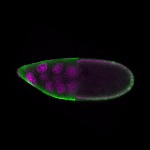
| stage10 | 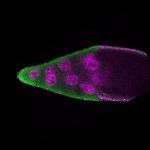 s s
 s s
 s s
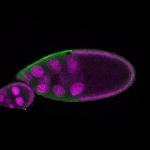 s s | somatic cells germline cells_ubiquitous nuclear foci follicle cells |
Quickview
| Probe source: | RE71517 (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | 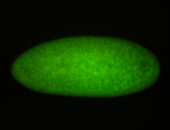
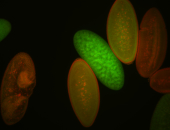
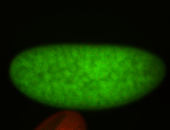
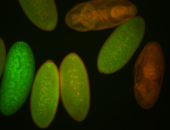
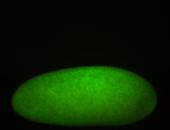
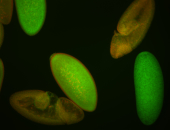 | Degraded Strong expression (1-20ms exp) Annotated by Hideki Stage 1-3 Degradation maternal transcripts Degraded partially stage 2 Ubiquitous (Unlocalized) Expressed Maternal |
| stage4-5 | 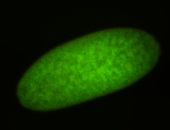
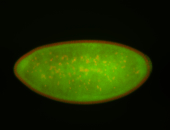
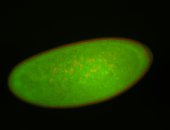 | Degradation maternal transcripts Stage 4-5 Ubiquitous (unlocalized) Expressed Degraded Degraded partially stage 4 Strong expression (1-20ms exp) Degraded partially stage 5 Maternal |
| stage6-7 | 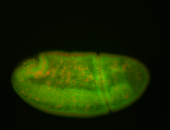
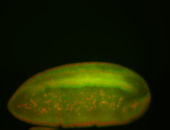
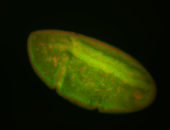 | Ubiquitous (Unlocalized) Medium expression (21-40ms exp) Stage 6-7 Expressed Maternal |
| stage8-9 | 
 | Ubiquitous (Unlocalized) Expressed Stage 8-9 Medium expression (21-40ms exp) Maternal |
| Probe source: | RE25591 (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| No images or annotation terms | ||
Quickview
| Probe source: | (best) RE71517 (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | cat_term:global_finito no staining | |
| stage4-6 | no staining list_4-6:no staining | |
| stage7-8 | no staining | |
| stage9-10 | no staining | |
| stage11-12 | ubiquitous foregut PR anal pad SA hindgut proper PR | |
| stage13-16 | brain foregut epipharynx dors epidermis visceral branch atrium vent nerve cord anal pad vent epidermis hindgut Malpighian tubule tip cell gonad midgut interstitial cell proventriculus outer layer midgut | |
| Probe source: | RE25591 (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | cat_term:global_finito no staining | |
| stage4-6 | no staining | |
| stage7-8 | no staining | |
| stage9-10 | no staining | |
| stage11-12 | ubiquitous foregut PR anal pad SA hindgut proper PR | |
| stage13-16 | central nervous sys brain foregut epipharynx atrium visceral branch dors epidermis anal pad vent epidermis vent nerve cord Malpighian tubule tip cell hindgut gonad proventriculus outer layer midgut midgut interstitial cell | |
| Probe source: | RE71517-dg (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | cat_term:global_finito no staining | |
| stage4-6 | no staining | |
| stage7-8 | no staining | |
| stage9-10 | no staining | |
| stage11-12 | ubiquitous foregut PR anal pad SA hindgut proper PR | |
| stage13-16 | brain foregut epipharynx visceral branch dors epidermis atrium vent epidermis anal pad vent nerve cord hindgut Malpighian tubule tip cell gonad midgut midgut interstitial cell proventriculus outer layer | |
| Probe source: | RE25591-dg (Flybase link CG32048 , FlyFish insitu , BDGP insitu new) | |
| No images or annotation terms | ||