Annotation summary for ref(2)P in insitu_ovaries_public
Legend: RNA signal: green, DNA staining: magenta
| Probe source: | GH06306-dg (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1 | 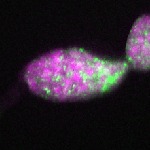 s s
 s s | |
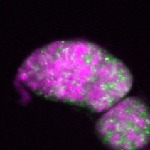
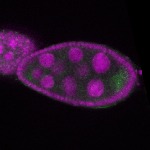
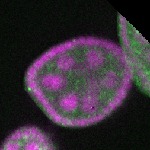
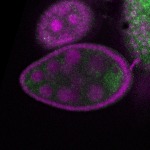
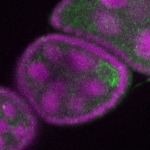
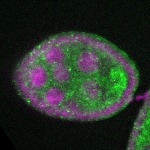
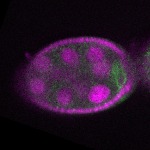
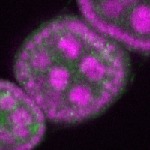
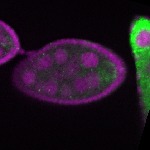
| stage2-7 | 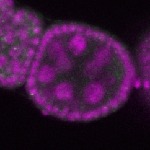 s s
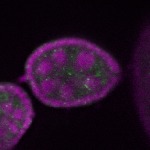 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
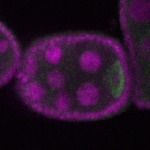 s s
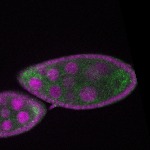 s s
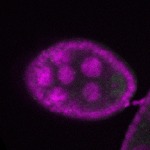 s s
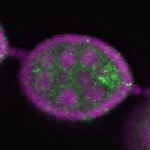 s s
 s s
 s s
 s s
 s s
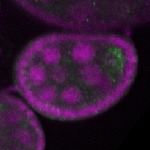 s s
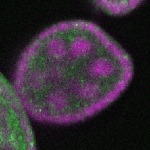 s s
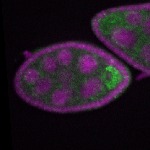 s s | enrichment_ubiquitous oocyte |
| stage8 | 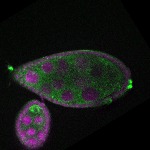 s s
 s s
 s s
 s s
 s s
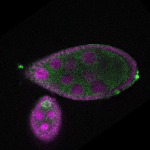 s s
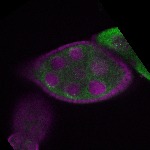 s s | ubiquitous signal |
| stage9 | 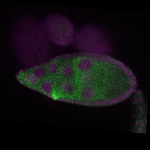 s s
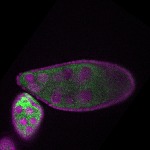 s s
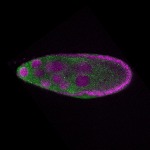 s s | cortical enrichment ubiquitous signal oocyte |
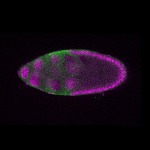
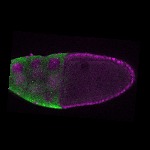
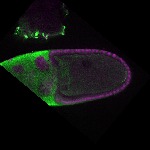
| stage10 | 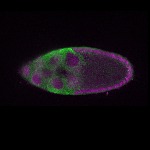 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
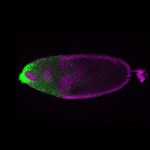 s s | oocyte cortical enrichment |
| Probe source: | RE24064 (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
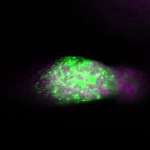
| stage1 | 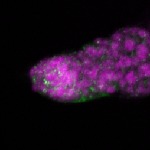 s s
 s s
 s s
 s s | |
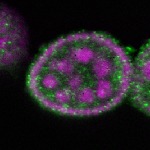
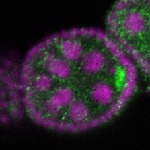
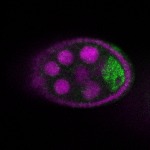
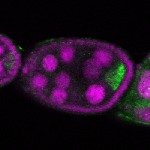
| stage2-7 |  s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s
 s s | oocyte enrichment_ubiquitous |
| stage8 |  s s
 s s
 s s | ubiquitous signal |
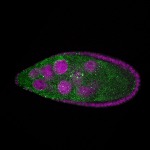
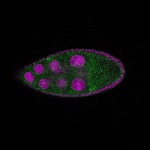
| stage9 | 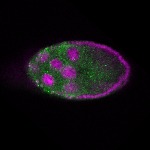 s s
 s s
 s s
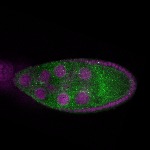 s s | ubiquitous signal |
| stage10 |  s s
 s s
 s s
 s s | ubiquitous signal |
| Probe source: | GH06306 (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| No images or annotation terms | ||
Annotation summary for ref(2)P in insitu_fish
Legend: RNA signal: green, DNA staining: magenta
| Probe source: | GH06306 (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | 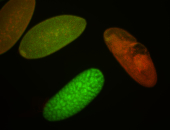
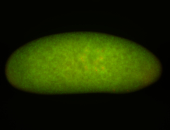
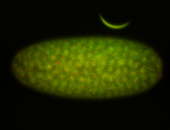
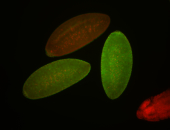
 | Localized Expressed Stage 1-3 Perinuclear around all nuclei Cell division apparatus Cytoplasmic foci Subcellular localization patterns Annotated by Ronit Embryonic localization patterns Perinuclear localization Strong expression (1-20ms exp) Nuclei associated localization Maternal |
| stage4-5 | 
 | Cytoplasmic foci Apical localization Subcellular localization patterns Degraded partially stage 5 Expressed Localized Discrete apical foci Pole cell enrichment Apical enrichment Degraded completely stage 5 Degraded Stage 4-5 Pole cell localization Embryonic localization patterns Maternal |
| stage6-7 | 
 | ant blastoderm nuclei Discrete apical foci Blastoderm nuclei Subset blastoderm nuclei Cytoplasmic foci Subcellular localization patterns Degraded completely stage 7 Degraded partially stage 7 Pole cell localization Stage 6-7 Expressed Localized Apical localization Zygotic starting stage 7 Zygotic Degraded Embryonic localization patterns Pole cell enrichment Apical enrichment Maternal |
| stage8-9 |  | Embryonic localization patterns Stage 8-9 Zygotic Subset blastoderm nuclei Blastoderm nuclei Localized Germ cell localization ant blastoderm nuclei Germ cell enrichment Expressed Subcellular localization patterns Cytoplasmic foci Maternal |
| Probe source: | RE24064 (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | 
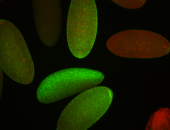

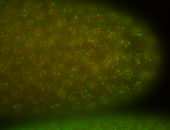
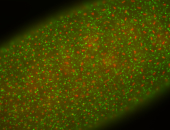
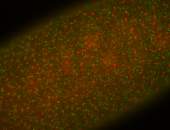
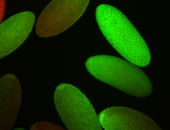
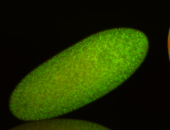
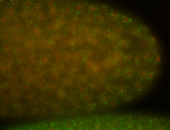
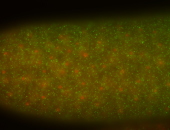

 | Cytoplasmic foci Subcellular localization patterns Perinuclear around all nuclei Stage 1-3 Expressed Localized Nuclei associated localization Medium expression (21-40ms exp) Perinuclear localization Annotated by Ronit Embryonic localization patterns Maternal |
| stage4-5 | 
 | Medium expression (21-40ms exp) Apical enrichment Nuclei-associated localization Embryonic localization patterns Apical localization Perinuclear around pole cell nuclei Expressed Localized Exclusionary patterns Basal exclusion Stage 4-5 Pole cell localization Perinuclear localization post localization Subcellular localization patterns Cytoplasmic foci Discrete apical foci Maternal |
| stage6-7 | 
 | Subset blastoderm nuclei Discrete apical foci Blastoderm nuclei ant blastoderm nuclei Random blastoderm nuclei Subcellular localization patterns Cytoplasmic foci post localization Perinuclear localization Pole cell localization Stage 6-7 Localized Expressed Perinuclear around pole cell nuclei Apical localization Embryonic localization patterns Zygotic Nuclei-associated localization Pole cell enrichment Medium expression (21-40ms exp) Apical enrichment |
| stage8-9 | 
 | Degraded partially stage 9 Subcellular localization patterns Germ cell nuclei Perinuclear around pole cell nuclei Expressed ant blastoderm nuclei Subset blastoderm nuclei Blastoderm nuclei Localized Nuclei-associated localization Weak expression (>41ms exp) Degraded Zygotic Stage 8-9 Perinuclear localization |
| stage10- | 
 | Blastoderm nuclei Localized Expressed Subcellular localization patterns Hindgut Zygotic post spiracles Stage 10- Specific hindgut area or cells Salivary glands - specific region Salivary glands vent nerve cord |
Annotation summary for ref(2)P in insitu
Legend: RNA signal: green, DNA staining: magenta
| Probe source: | RE24064 (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | maternal | |
| stage4-6 | ubiquitous list_4-6:specific | |
| stage7-8 | ubiquitous pole cell | |
| stage9-10 | ubiquitous procephalic ectoderm PR trunk mesoderm PR head mesoderm PR ant endoderm PR post endoderm PR germ cell | |
| stage11-12 | ubiquitous ant midgut PR post midgut PR germ cell | |
| stage13-16 | ubiquitous cat_term:global_finito hindgut vent midline crystal cell midgut | |
| Probe source: | GH06306 (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| stage | images | Annotation |
| stage1-3 | maternal | |
| stage4-6 | list_4-6:specific cellular blastoderm | |
| stage7-8 | list_7-8:specific trunk mesoderm PR ant endoderm A post endoderm PR pole cell | |
| stage9-10 | trunk mesoderm PR ant endoderm PR | |
| stage11-12 | germ cell | |
| stage13-16 | germ cell | |
| Probe source: | GH06306-dg (Flybase link CG10360 , FlyFish insitu , BDGP insitu new) | |
| No images or annotation terms | ||