Welcome to DOT, the Dresden Ovary Table - the genome-wide resource about gene expression and RNA localization in fly ovaries.
The DOT presents visual 3D microscopy-based information about RNA localization for more then 1700 genes in the fruit fly adult ovary at all time-points of oogenesis. Altogether, the database contains more then 8000 in situ experiments documenting almost 7000 genes with 32,000 image stacks and over 50,000 exemplary 2D images. Additionally, RNA seq and 3'UTR seq data covering relevant portions of oogenesis and early embryogenesis are presented for all genes in the genome.
All information is Open Access, searchable and can be downloaded and re-used in your own studies. We have developed several methods that give best results for fluorescent in situ staining and immuno-staining, and we also established a protocol for rapid mass-isolation of egg-chambers from flies.
Currently the core team of the DOT are Helena Jambor a postdoc and Pavel Tomancak who are interested in understanding cytoplasmic regulation at a system biology level. The publication associated with the DOT is available here: Systematic imaging reveals features and changing localization of mRNAs in Drosophila development.
For any enquiries, comments please get in touch!
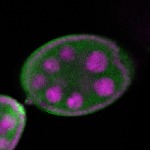 | 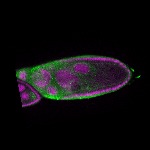 | 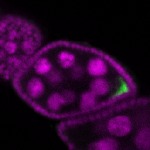 | 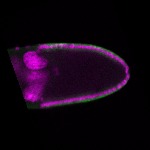 | 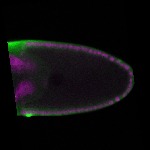 | 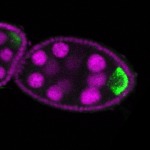 | 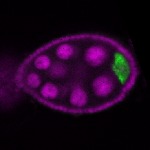 |